Product
A modern data & compute
agent for biotech R&D.
Hundreds of biotechs use Latch to make data analysis faster, cheaper, more accessible, and instantly accelerate their R&D milestones.
Make scientific decisions faster than ever.
Centralize and standardize all your data and computation into a single source of truth.
Latch Data
A storage solution for modern R&D.
Connect your instruments, workflows, and reports into your own private workspace secured by Amazon s3.
“The increased accessibility to data has led us to generate new therapeutic targets.”
- CSO, RNA Therapeutics Biotech
Enterprise data security
Secure and private in your own cloud environment.
Keep ownership and privacy over your data on Amazon s3 with 99.9999999999% (11 9s) of data durability. Latch helps you save months and become compliant on day one with the latest regulations, standards, and industry frameworks such as SOC-2 Type II and HIPAA. Read more at trust.latch.bio.

HIPAA

SOC-II
Unlimited storage
Store any data, any size, from anywhere.

A simple and accessible way to see every piece of data your team generates. Import from Benchling, AWS, DropBox, GCP, or Azure.

Fine-grained user permissions
Track and trace every single change.
Keep an audit trail of who did what, and when. Accelerate daily tasks and your FDA IND proposals alike.


Invite unlimited users.
Manage your users' access and permissions.
Manage members and workspaces under your organization. Invite unlimited users with fine-grained access controls.



Loved by wet and dry lab.
Give scientists access to data, quickly
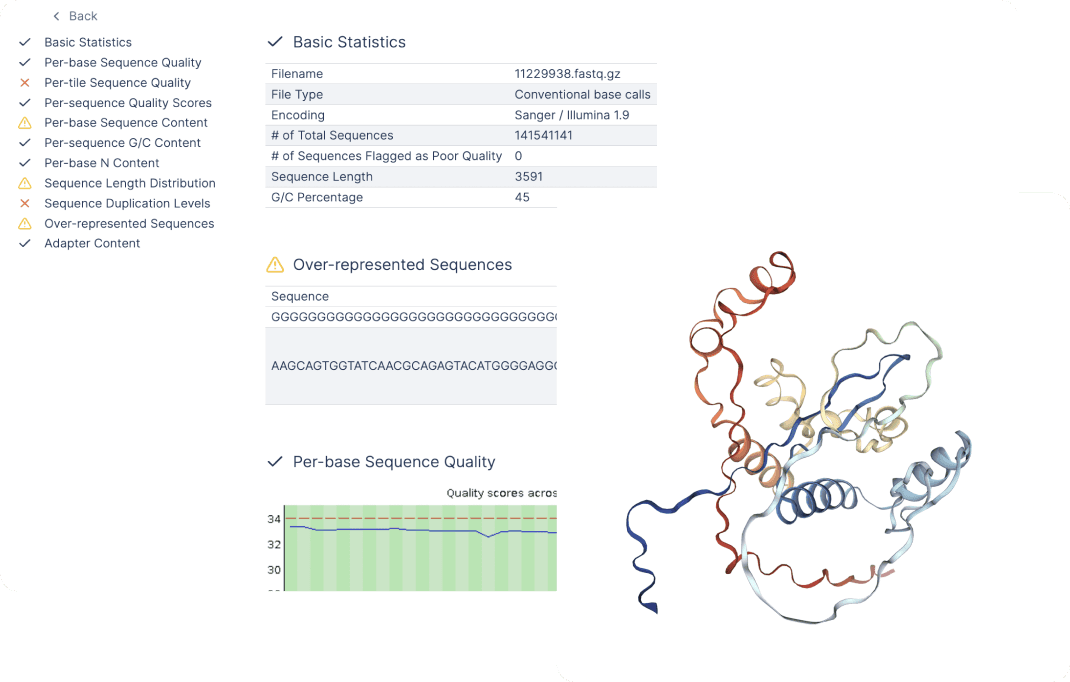
NGS files (FastQ, BAM, SAM, VCF, PDB) have default visualizations, such as FastQC, IGV, and Protein Viewer built on top, so you can inspect key insights in seconds, without having to ever leave the Latch platform.
Latch Registry
An integrated database to harmonize metadata
Never lose track of mission-critical metadata
“With Registry, you can tie all of the sequencing data, strain images, and metadata together in one place. It makes analysis much more comprehensive and enables both scientists and bioinformatics team interface with the same images, sequencing files, and metadata in a table-like format.”
Chris Baldock, Chief Data Officer, Metagen
A collaborative database
A shared database for the wet and dry lab
A familiar spreadsheet-like interface that is easy to edit and reference files stored in Latch Data. Embed images or links to reports in the table for quick interpretation of biological data and create parent-child relationships between tables.

Sequencing File Organization at Scale
Organize raw instrument files with metadata
Catalog and link files on Latch Data with metadata in a table-like format which allows you to designate strong types to columns in tables to enforce standardization. And import bulk data via a spreadsheet like Excel or CSV

Plugs into your workflows
Connect code and pipelines to registry
Connect your pipelines to read and write to registry for better traceability. Use APIs to easily query for patient cohorts or key information.

Findable
Search and find data instantly
Effortlessly search and filter Registry tables to retrieve past data based on specific column and cell values.

Developer-friendly
Update data with Python API
Registry Python API allows for CRUD (create, read, update, and delete) operations on Registry tables. Read and write to Registry tables from a workflow or notebook environment
...
file_path = Path(file.local_path)
file_size = os.Path(file_path).st_size
tbl = Table(id="1234")
with tbl.update() as updater:
tbl.upsert_record(
record_name,
{
"File": file,
"Size": file_size,
}
)
...Latch Workflow Manager
A Workflow Orchestration Platform for Scalable Analyses
Upload any bioinformatics workflow with flexibility. Empower scientists to self-serve workflows.
“Our developer uploaded workflows to Latch that can analyze 100 curves at a time. And all scientists need to do is click a button on Latch. This saves an enormous amount of time, probably a day of work per scientist, 20 days a month of labor.”
Eszter N. Tóth, PhD, Senior Research Scientist, Etcembly
Get a user-friendly interface. No extra work.
Auto-generation of workflow interface
Latch SDK is a flexible Python SDK that enables easy portability from existing frameworks and languages to Latch.
Define tasks using pure Python functions and leverage the subprocess module to spawn child processes that execute scripts in any language.
@workflow
def my_workflow(
...
read: LatchFile,
amplicon_sequence: List[String],
quantification_window_center: Union[None, int] = -3,
...
):
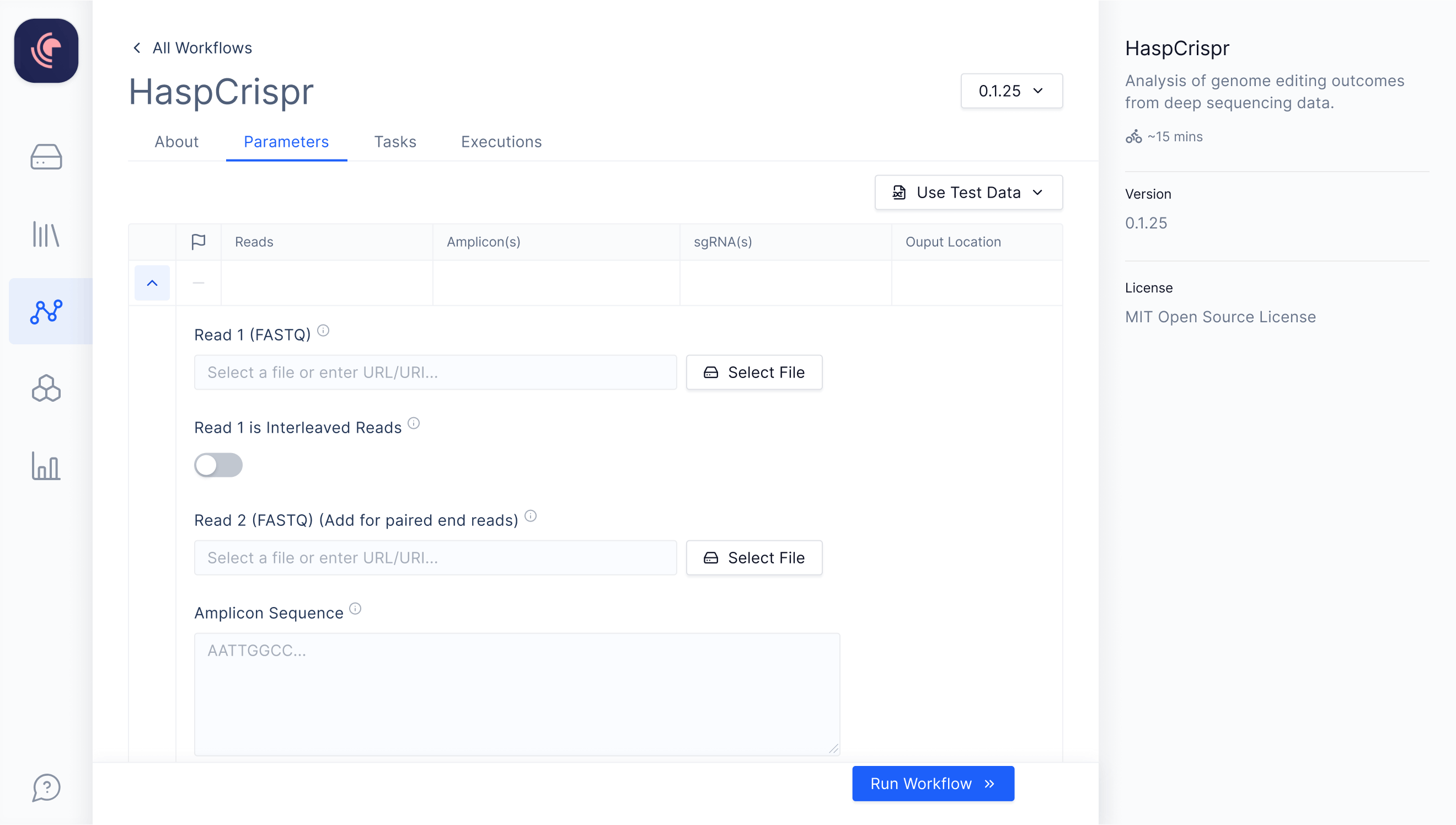
...Each parameter can be configured by altering its corresponding component (swapping a number field with radio options for common integer values), adding documentation (such as labels and help tooltips), and by changing its order and grouping on the page.
@workflow
def my_workflow(
...
read: LatchFile,
amplicon_sequence: List[String],
quantification_window_center: Union[None, int] = -3,
...
):
...
read:
A fastq file of sequenced reads
__metadata__:
display_name: Reads
appearance:
batch_table_column: true
detail: (.fastq, .fastq.gz)
placeholder: Select a file or input URL/URI...
amplicon_sequence:
The amplicon sequence(s) used for the experiment
__metadata__:
display_name: Amplicon Sequence
appearance:
placeholder: AATTGGCC...
quantification_window_center:
Center of quantification window to use within respect to the 3' end of the provided sgRNA sequence.__metadata__:
display_name: Quantification Window Center
appearance:
multiselect:
options:
- {name: Cas9, value: -3}
- {name: Cpfl1, value: 1}
- {name: Base Editors, value: -10}
allow_custom: true
Scalability
Specify arbitrary resource your workflow needs
Use single-line python task decorators to easily define resources available at runtime.
The framework starts at 2 CPUs and 4 GBs of memory and goes all the way to 31 CPUs, 120 GBs of memory and 1 GPU (24 GBs of VRAM, 9,216 CUDA cores) to easily handle all processing needs.
@latch.medium_gpu_task
def cas_offinder(
...
):
@latch.large_task
def nf_core_rnaseq(
...
):
...Latch Pods
Scalable, on-demand cloud computers
Productivity and flexibility of a personal computer. Scale of the cloud.
“Our company is extremely exploratory, I wanted something extremely flexible. I was looking literally for an extension of my personal computer. I needed to be able to write my own code, the way that I know how to do it today, and to connect that to seamlessly memory and compute.”
Jonathan Hsu, Chief Technology Officer, Gensaic
Scalable resources on-demand
Start large cloud computers instantly
It’s that simple - select up to 93 CPU cores, 8 GPUs, 730 GiB RAM, and 16000 GB of storage and click “Launch”

Accessible notebook environment
Run analyses in notebook environments
Built-in Jupyter Lab and RStudio to support Python and R analyses & bash terminals for command-line operations.
...or use it with your favorite IDE — connect to Pods via SSH from your local computers and integrate with a familiar IDE (VSCode, PyCharm, and more) .
Reproducibility at its core
Save, share, and reuse compute environments
Once your Pod is set up with dependencies, scripts, and analyses, you can save a snapshot of your Pod into a Pod Template.
All members in the same team workspace can start a new pod from a template. Make pod templates “Public” to share it with the world, or re-use others.
Be in charge of what you pay
Save cost with auto-shutoff
Fine-grained options to auto shutoff a Pod if there is no network activity to save cost

World-class Customer Support
More than just a platform – an analysis partner
Don’t take our word for it.
The thing with Latch is the customer service is brilliant. No matter how good a platform is, if the customer service is not good, you’re not going to use it. Latch responds right away to literally any question. It’s quite insane actually.”
Eszter Toth - Group leader of Genomics, Etcembly
The people behind Latch and the responsiveness is awesome. I have been very impressed with how fast they respond to us. It feels like we’ve got this entire LatchBio team at our fingertips to solve the problems we’re not equipped to solve. That definitely exceeded expectations.”
Jonathan Hsu - Chief Technical Officer, Gensaic
The support and onboarding we received from latch has been way beyond our expectations. The platform has been a great solution for our complex spatial biology data.”
Colin Ng - Vice President of Business Development, AtlasXomics
How we work with Partners
We take your scientific journey seriously.
With Latch, we map out your entire end-to-end drug discovery campaign. Our engineering team acts as an extension to your company and provides support to migrate all data, workflows, and notebooks upfront. For early-stage companies, our in-house bioinformaticians provide consulting services to get your first analyses up and running as fast as possible. That way, you have more time doing life-saving science, and less time worrying about cloud infrastructure set up.
Accelerate biological insights now
Request a Demo
Access the leading data platform for biology.
- The best cloud platform for processing of biological data
- Save over 72% vs. AWS, GCP, and Azure
- Trusted by more than 4,000 scientists
- Built for biological data
- Start today