Latch Workflows
Run Nextflow Based Pipelines at Scale
Integrate workflows built with Nextflow on the platform hundreds of biotechs use to make data analysis faster, cheaper, more accessible, and instantly accelerate their R&D milestones.
An optimized platform for workflows built with Nextflow
Latch addresses infrastructure challenges for workflows built with Nextflow and facilitates their delivery to scientists.
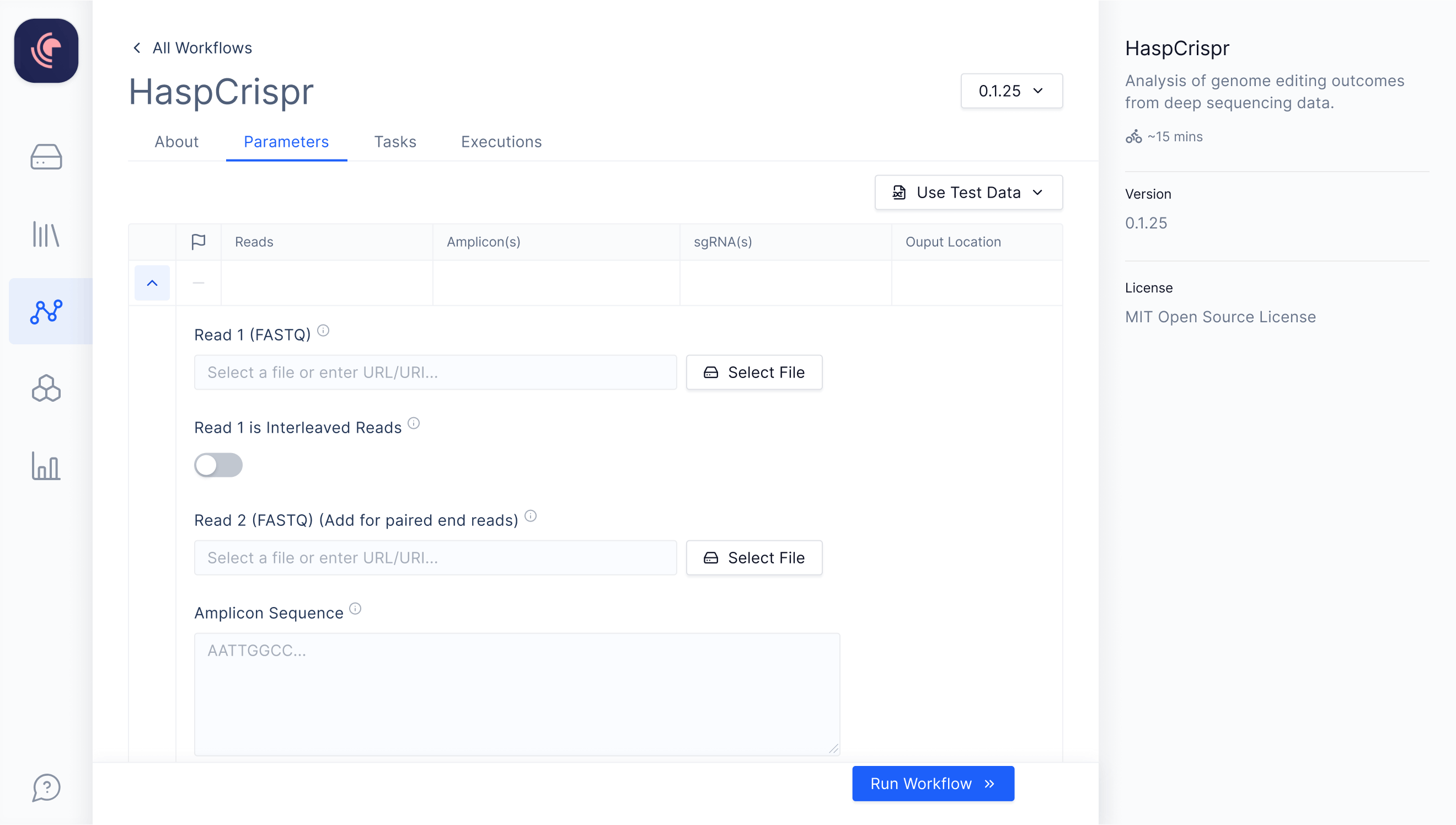
User-friendly, Type-safe Interfaces
Latch integrates with workflows built with Nextflow, enabling developers to create user-friendly graphical interfaces accessible to wet lab scientists.
No changes required to existing Nextflow code
Customizable interface via an additional Python file
Bring data from anywhere - AWS, GCP, SRA, BaseSpace, and more.
Downstream analysis tooling
With Latch, scientists can transform raw output files from workflows built with Nextflow into interactive visualizations, facilitating critical scientific decisions.
Results to Plots
Downstream analyses in Pods
Native visualizations (FastQC, CELLxGENE) for common files
Built for Developers.
Latch offers managed cloud infrastructure for executing, debugging, and analyzing workflows built with Nextflow.
It uses a customized Nextflow Kubernetes plugin to run processes in containers within Latch’s Kubernetes cluster, which allows developers to use existing Nextflow projects with minimal code alterations.
Intermediate outputs are shared via AWS EFS, and final outputs can be uploaded to Latch Data using a custom plugin.
The Latch Console features a Directed Acyclic Graph (DAG) for monitoring progress, checking logs, and analyzing process costs and runtime. Developers also have real-time access to the container for debugging purposes.
Deploy existing Nextflow workflows with minimal changes
Migrate workflows built with Nextflow to LatchBio in three simple steps while maintaining full portability.
Boost pipeline throughput and reduce cost via a shared filesystem
Enhance performance of workflows built with Nextflow by using a shared filesystem for efficient data handling.
Optimize workflows with real-time per process cost and runtime reporting
Obtain real-time insights on costs and runtime for each process in workflows built with Nextflow to optimize efficiency.
Scalable and reliable for large workloads
Manage large-scale workflows built with Nextflow using LatchBio’s scalable, high-performance infrastructure.
Retry from failed tasks
Quickly modify and relaunch workflows built with Nextflow from the point of failure without re-entering parameters or restarting the entire workflow.
Designed for reproducibility with GitHub integration
Ensure reproducibility of workflows built with Nextflow through automatic linking of versions to corresponding GitHub commits.
How it works
Upload your own
Install Latch & Clone your repositories containing workflows built with Nextflow
$pip install latch$git clone https://github.com/nf-core/rnaseq$cd rnaseqDefine metadata and workflow graphical interface
$latch generate-metadata nextflow_schema.json --nextflowMetadata files:
latch_metadata/__init__.py
latch_metadata/parameters.py
Register the workflow to Latch
$latch login$latch register . --nf-script main.nf --nf-execution-profile docker– and that’s it. You just uploaded the nf-core/rnaseq pipeline to Latch!
Or access the nf-core library on Latch
nf-core/rnaseq
nf-core/sarek
nf-core/scrnaseq
nf-core/methylseq
nf-core/mag
nf-core/atacseq
nf-core/nanoseq
nf-core/ampliseqs
nf-core/cutandrun
nf-core/bacass
Each pipeline on Latch has a user-friendly interface, a graphical, error-validated sample sheet component to fill out pipeline inputs, and a directed acyclic graph (DAG) to view process and sample-specific errors.
For Bulk RNA-seq, ATAC-seq, and Methyl-seq, Latch also offers interactive plotting dashboards that directly ingest NF-core outputs and produce publication-ready figures.
Features
Performance Optimization
Easily view and compare results, cost, and performance of different executions
Graphical workflow interface
Easily view and compare results, cost, and performance of different executions
Integration with Latch Registry
Developers can extend their existing workflows built with Nextflow to read from Latch Registry, a user-friendly, error-validated sample sheet input system.
Debugging
Avoid failed workflows caused by discrepancies between running code locally and in the cloud.
Latch SDK comes with the latch develop command, which drops you into an interactive shell where you can run your code and inspect the environment before registering and executing the entire workflow in the cloud to debug environments and logical issues.
Relaunching from failed tasks
The integration with Nextflow uses caching to save intermediate workflow results, allowing for relaunch without full re-execution.
Latch also exposes storage expiration hours as a configurable parameter to users to determine how long the cache will persist after a failure.
Logging
To facilitate debugging, a DAG of processes is generated in the Latch console to enable developers to easily track progress, view logs, and analyze cost and runtime of each process execution.
EFS integration
Intermediate outputs are communicated between processes via AWS EFS and published outputs can be uploaded directly to Latch Data via a custom filesystem plugin. The work directory is automatically cleaned up after workflow completion, saving costs and eliminating the hassle of manually cleaning up resources
Ready-to-use NF-core pipelines
Latch has top 10 ready-to-launch NF-core workflows with user-friendly interfaces, documentation, and GitHub repositories to help you get started.
Sharing of pipelines
There’s no seat limit on Latch. Everyone with a Latch account can create any number of workspaces and add unlimited team members to each workspace. As an Admin, user can also distribute versions of their pipelines built with Nextflow to multiple workspaces.
Private registries
Latch Secrets allow Nextflow processes on Latch to securely use containers images hosted in users’ private registries (GitLab, AWS ECR, Azure container registry, and more)
Integration with GitHub
Workflows on Latch can be versioned using Git. If the SDK detects that the project directory is a Git repository, it will append the first six digits of the latest commit hash to the workflow version.
LatchBio, LATCH, and the Arrow Logo are trademarks of LatchBio, Inc. All other trademarks are the property of their respective owners.
Request a Demo
Access the leading data platform for biology.
- The best cloud platform for processing of biological data
- Save over 72% vs. AWS, GCP, and Azure
- Trusted by more than 4,000 scientists
- Built for biological data
- Start today