TL;DR
Metagen implemented LatchBio to automate processing metabarcoding data and share the results with their scientists and collaborators.
Using Latch, Mategan Achieved:
1 Full Time Engineer Saved
Adopting Latch removed the need to manage AWS infrastructure, which would have otherwise required hiring a full-time engineer.
10x faster NGS Analysis
Using Latch, running NGS analysis went from several hours to 10 minutes.
Made data accessible to their 8 scientists
Metagen's 8 scientists can now query, access, and analyze data themselves, removing bottlenecks at the computational level
Metagen’s story
Metagen is an Australian owned biotechnology company that works with Australian growers to optimize crop productivity, profitability, and sustainability. By using advanced microbial technology, they offer a new generation in agricultural analytics, resulting in immense productivity gains, massive cost savings, and improved soil health for their partners.
For their customers, Metagen collects:
- Strain Collection Images and Crop Metadata: Collected via Metagen’s phone applications, growers send photos and data about crop health to Metagen’s scientific team to help them remotely diagnose disease and monitor treatment progression
- rRNA Marker gene Sequencing Data: Once growers send samples of their crops to the lab and fill in the relevant metadata, the Metagen scientific team will analyze bacteria and eukaryota rRNA marker gene sequencing using the DADA2 pipeline to understand the microbial species present. They then compare these species to past samples to understand which microbes are driving disease progression and impacting productivity
- Functional Gene Prediction: After taking in in the outputs of DADA2 (ASV Sheets), their functional genomics pipeline outputs the number and roles of genes present in any given sample. This enables their team to classify the effects that specific species of fungi and bacteria will have on crops.
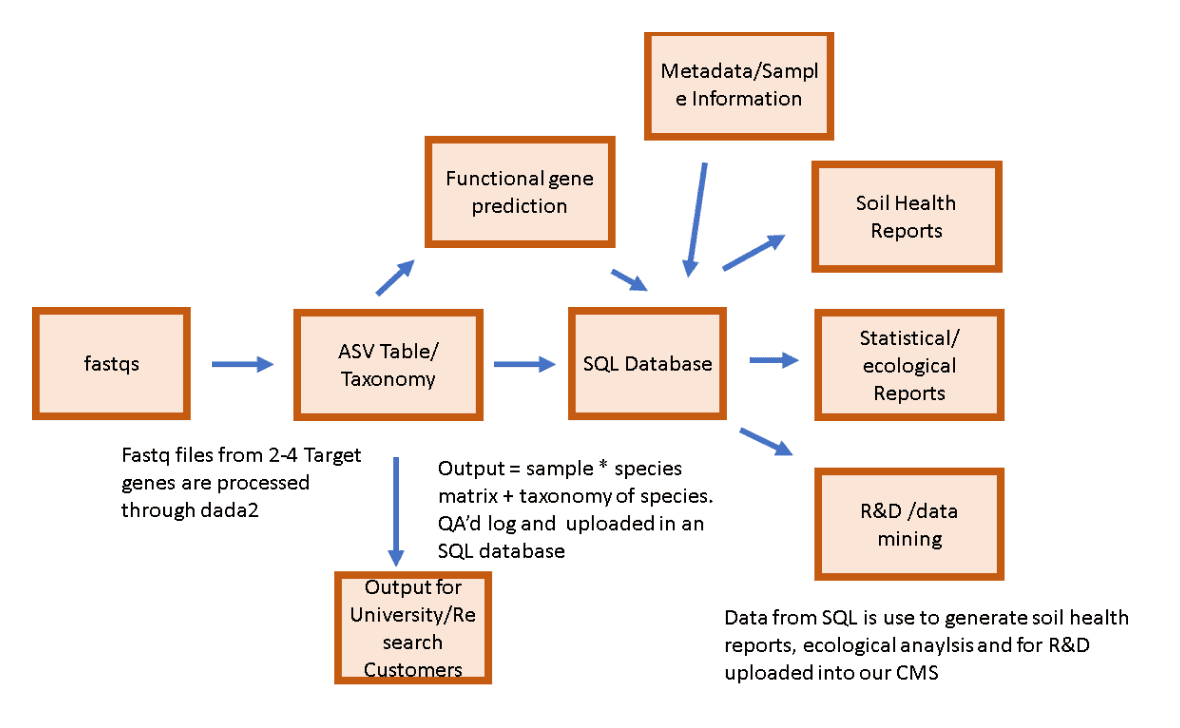
Then processes all data and provides these deliverables:
- Soil and Health Reports: These reports are served directly to farmers to help them understand their current soil health, how it is impacting crop productivity, and how Metagen’s treatments and offerings would be valuable to them.
- Ecological Reports: These reports help Metagen’s team, research collaborators, and farmers understand which species are enriched/depleted based on their various treatments.
- R&D/Data Mining: These tools are used to identify underlying mechanisms driving changes in microbial community structure to improve Metagen’s understanding of microbial communities on soil and plant health.
Challenges in Delivering Analysis to Clients at Scale
CXO Time Wasted Manually Configuring AWS
Chris, Metagen’s Director, would spend hours manually spinning up, tearing down, and configuring multiple AWS instances of varying sizes to run workflows, requiring him to pipe data in between each step and check AWS continuously for hours. This delayed him getting results to the rest of the team for further analysis
The sequential bioinformatics workflows often use different types of software so you have to wait for one process to finish, manually start another instance, and pipe the data over while checking for errors. Tying everything together was the real bottleneck for the rest of of my team.”
Slow Turn-Around Times for Customer Data
Metagen serves dozens of Australian growers with bespoke needs and truncated timelines due to their growing seasons. Because Chris is the sole computational person at Metagen and has other responsibilities (including attending conferences, visiting the fields, or assisting in the lab), time-sensitive customer analysis is frequently delayed.
A lot of the metabarcoding runs we do are for commercial testing, so we need to turn those around as quickly as possible. If I’m trying to do something else, I can’t be running the pipeline. If there are conferences, or meetings, or other duties I need to fulfill, those have to take a backseat and that slows the whole company down.”
Difficulty Linking Sequencing Data with Metadata
Metagen grows and visually analyzes hundreds of images of strains cultured in the lab to identify bacteria and fungi that commonly grow on patient crops. This process is extremely manual, and does not enable them to link the photos with the genetic sequences of the samples themselves, which would greatly increase their ability to identify species.
We have a strain collection of bacteria and fungi stored in an Excel format that our scientists have to manually search every time we want to cross reference or share results with growers or agronomists in the field. Not being able to link the large sequencing files directly to the images and other metadata is time consuming and is organizationally challenging.”
Ambiguous Error Handling
Chris would frequently run into errors with his analysis when working with AWS directly, and it would be challenging to find and identify the errors within the runs.
Given the amount of data that we’re generating you can’t just use local machines, so we were using AWS instances to run workflows. But there’s a fair bit of manual handling of the scripts and spinning up the instances.
Solution
Latch is a biological cloud where organizations can store, process, analyze, and visualize multi-omics data. Metagen believed that Latch could address their challenges with doing reproducible analysis at scale for its clients while eliminating the need to maintain their own cloud infrastructure.
Metagen engaged in a free 1-week trial with Latch where our teams collaborated to successfully migrate all of their workflows, data, and scientists onto the platform.
For anyone on the fence about trialing Latch, go for it! It might seem daunting at first, but it’s easy to use, and they have a resource of technical expertise there, ready to go.”

Outcomes
Accelerated the Time to Run Routine Analysis from 3 hours to 3 minutes
Using LatchBio’s user friendly front-end interface, Chris can launch hundreds of analytical jobs at once in a matter of minutes. All he has to do is select his files, input the parameters, and click “Launch Workflow”. He no longer has to manually spin up and tear down instances, install scripts, or move data manually, saving him a massive amount of time each day.
Increased Data Accessibility and Ownership for Scientists
Using Latch Registry, Metagen’s team can link files of any size and type with their associated metadata. This enables their scientists to explore their strain collections of bacteria and fungi alongside their sequencing files in a user friendly way, freeing Metagen’s computational team from manually searching Excel sheets and enabling them to run more metagenomic analysis than before.
Reduced Engineering Load by 1 FTE on the Computational Team
Metagen no longer needs to manage their AWS infrastructure and serve results to scientists. This frees up precious engineering time to work on other high-priority projects.
Increased transparency with strict versioning, containerization, and accessible logs
Once migrated to LatchBio using the LatchBio SDK, all of Metagen’s bioinformatics workflows are automatically versioned and containerized, ensuring their reproducibility. Execution logs are also prominently displayed for each workflow run, enabling a much quicker debugging process.

Latch has absolutely helped us expand what we can do scientifically and computationally.
Latch is the bridge between the wet lab and in-silico lab. It’s your direct feed from translating what you do in the lab on a day-to-day basis onto a unified platform to analyze trends from experiments. It’s a one-stop shop - it has everything you need.”


