TL;DR
Funga uses Latch to store thousands of samples of fungal sequencing data, provision cloud resources to process this data in minutes with custom workflows, and version all of their pipelines and analysis.
Using Latch, Funga achieved:
Accelerated Cloud Deployment
Latch enabled their team to instantaneously deploy workflows to the cloud with no DevOps experience required
Centralized Pipelines and Analysis
With Latch all of their analysis tools and results were in one place which allowed for real-time collaboration between their wet and dry-lab teams
Guaranteed Reproducibility and Traceability
Latch automates tracking and versioning for every workflow developed ensuring everything is reproducible
Funga's story
Funga harnesses forest fungal networks to address the climate crisis and improve microbial biodiversity. By combining modern DNA sequencing and machine learning technology with breakthrough research on the forest microbiome, Funga can put native and biodiverse communities of fungi in the right place to accelerate tree growth, wood production, and carbon removal in both forestry and restoration contexts.
These microscopic organisms have profound effects on forest growth and carbon capture that until now have been overlooked as a way to accelerate natural climate solutions while also restoring essential microbial biodiversity to our soils.”
- Colin Averill, Founder & CSO
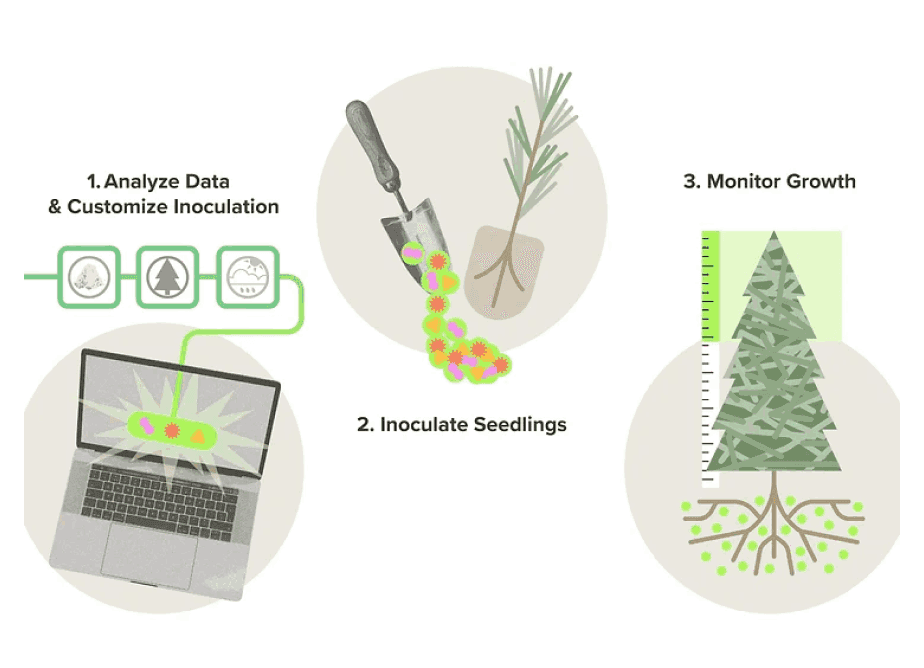
To accomplish this, the Funga team collects soil samples from forests around the world in an effort to understand their fungal populations. After sequencing their samples, they can then perform machine learning using the sequencing data and forest productivity metrics to identify how fungi affect tree growth, tree health and carbon capture.
Performing analysis on thousands of samples, however, requires:
- Scalable cloud infrastructure for computation/storage
- Pipeline versioning since this study will be taking place over the next several years and use several packages for analysis
- A collaborative environment to host and share analysis
Challenges
Lack of Cloud Infrastructure Knowledge
Because Funga’s pipeline contains multiple workflows requiring vastly differing amounts of resources (sequence processing vs taxonomic classification vs machine learning), it could have taken months of time and cost $100,000s in hiring costs to learn to provision the required cloud resources to run these pipelines on their own infrastructure.
“Using Latch, the ability to define something as a small task, medium task, large task […] makes it a lot easier than if we were to use AWS or Google Cloud Services.”
Decentralized Analysis
Because Funga is fully remote, they understood the need for a cloud based system to share, organize, and track data across their entire team.
The popular academic approach is to use multiple RScripts saved on multiple local machines, then store sequencing files on a hard drive…nothing like a unified pipeline.”
Unreproducible Pipelines
Because Funga’s packages are being constantly updated, pipelines can quickly become deprecated and require significant maintenance. On top of that, it’s easy to lose track of versions and your analysis is no longer reproducible, making it impossible to analyze data from the past to do accurate comparisons.
We use a bunch of different packages, so CutAdapt is going to be updated, Dada2 is going to be updated, and then for analysis we use 4 or 5 different packages, and there’s no real version tracking….it’s a huge problem.”
Solution
The LatchBio SDK is a framework to create new bioinformatics workflows, allowing users to provision cloud infrastructure, automatically generate frontend interfaces, and gain reproducible + versioned pipelines - all from a handful of python functions, completely bypassing the need to interface with a provider like AWS. In one week, Funga’s team successfully leveraged the SDK to help them instantly gain access to scalable cloud infrastructure and centralize all of their analysis.
After sequencing, thousands of Funga’s microbiome sequences are stored on the LatchBio platform, where their entire team can access them. Funga processes these sequences using the CutAdapt pipeline (publicly available on Latch) to trim the primers from the reads. They then utilize the LatchBio SDK to upload and run the Dada2 workflow, which infers exact amplicon sequence variants (ASVs) from high-throughput amplicon sequencing data (# of reads per bacteria/sequence type) and assigns taxonomy(classifying the microbes by species using the UNITE taxonomic database). With this bioinformatic data, Funga can identify exactly what fungi are where.
Challenges
Cloud Deployment
Funga was able to instantaneously deploy workflows to the cloud without ever needing to interface with AWS or have any prior DevOps knowledge.
Using Latch, the ability to define something as a small task, medium task, large task […] makes it a lot easier than if we were to use AWS or Google Cloud Services.”
Guaranteed Reproducibility and Traceability
All pipelines uploaded to Latch are automatically versioned and containerized, enabling Funga to track each and every pipeline they publish and simplify the development process.
We use a bunch of different packages, so CutAdapt is going to be updated, Dada2 is going to be updated, and then for analysis we use 4 or 5 different packages, and there’s no real version tracking….it’s a huge problem.”
Centralization of Pipelines and Analysis
All data and analysis was stored on the LatchBio platform, enabling users to collaborate and share their work in real time.
The popular academic approach is to use multiple RScripts saved on multiple local machines, then store sequencing files on a hard drive […] nothing like a unified pipeline.”
Conclusion
Before Latch, Funga had to set up this pipeline and computing locally, host it, troubleshoot it, and try and communicate results between the scientist and bioinformatician inefficiently.
Now their bioinformatics team can put Latch on their benchtop, drop in their sequencing data, and launch their analysis, safely scaling their operation and saving months on infrastructure set up. What was previously a bottleneck is now an automated solution enabling the team to design incredible workflows that accelerate Funga's ability to process large volumes of sequencing data.
LatchBio’s interface was intuitive and the documentation was easy to understand, even though I don’t have tons of coding experience.
They saved me tons of time from having to set up my own cloud infrastructure and learn how to use AWS. With them, I was able to access a customized, production grade bioinformatics environment in matter of weeks.”
- Caylon Yates, Microbiome Scientist


