TL;DR
AtlasXomics uses microfluidics and Next Generation Sequencing (NGS) to create spatial tissue maps that highlight the mechanisms of disease for researchers, but massive data challenges (unique workflows, enormous datasets, cloud infrastructure) made it difficult for scientists to easily access meaningful insights.
Using Latch, AtlasXomics:
Reduced time to insight by 96%
"Before we started working with Latch, it would take months to the bioinformatics work we had to do. And now it can be as little as two days." -CEO, Ken Wang
Made analysis tools accessible to biologists
"It’s made a huge difference to our own services, and more importantly it’s made a huge difference for our customers when they want to do the analysis on their own." -CEO, Ken Wang
Fostered wet lab <> dry lab collaboration
Scientists began investigating data and asking questions in entirely new ways, while the dry lab saved time on routine analyses.
AtlasXomics’ Story
AtlasXomics is offering a spatial biology platform using microfluidics and NGS to create spatial tissue maps that highlight the mechanisms of disease for researchers.
Their breakthrough technology was invented by the research group of Dr. Rong Fan, Professor of Biomedical Engineering at Yale, with high-impact publications in Cell, Nature, and Science. These pioneering studies showed comprehensive, unbiased epigenome, transcriptome, and proteome profiling in tissue at cellular resolution.
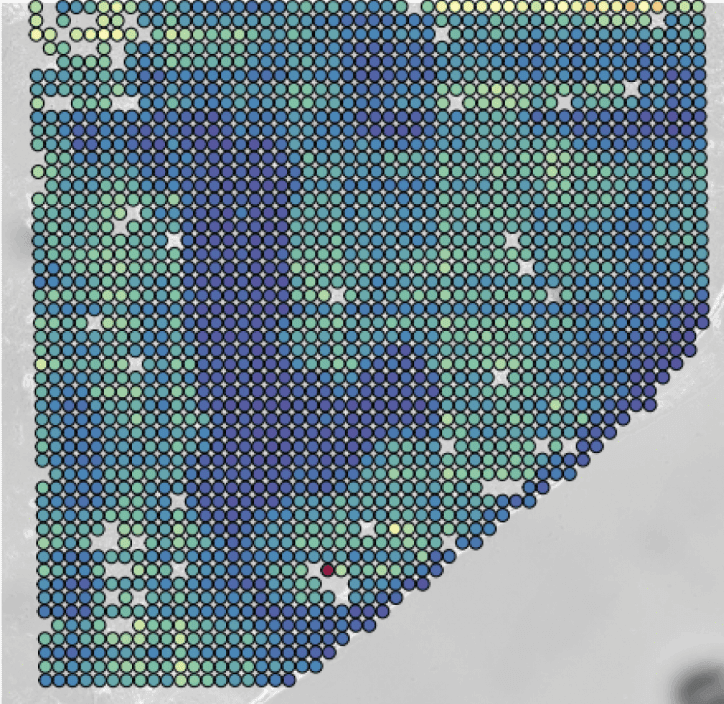
Maps like these reveal the underlying mechanisms that drive complex human diseases including cancer, autoimmunity, and cardiovascular disease. Generated by an ingenious combination of microfluidics chip design and NGS.
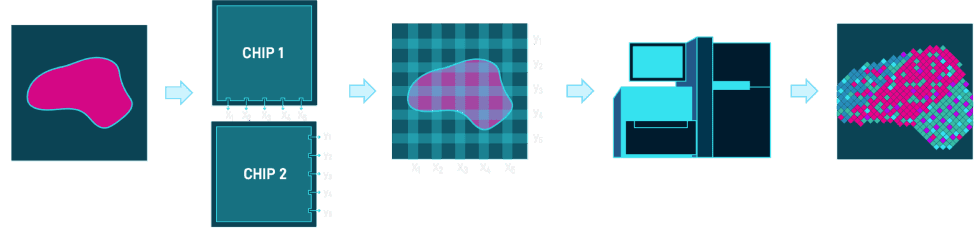
Figure 1: AtlasXomics’ method to reconstruct a spatial map of activity from raw tissue
The team of driven researchers are now focused on translating this technology to enable scientists across the world to easily unlock these insights. They care deeply about access:
We are trying to make spatial biology easy for everyone. Our long-term vision is to provide an accessible solution for labs to utilize spatial biology to answer their biological questions.”
The team began carrying out this mission with spatial epigenomics: a method of showing how gene expression is tuned by epigenetic elements.
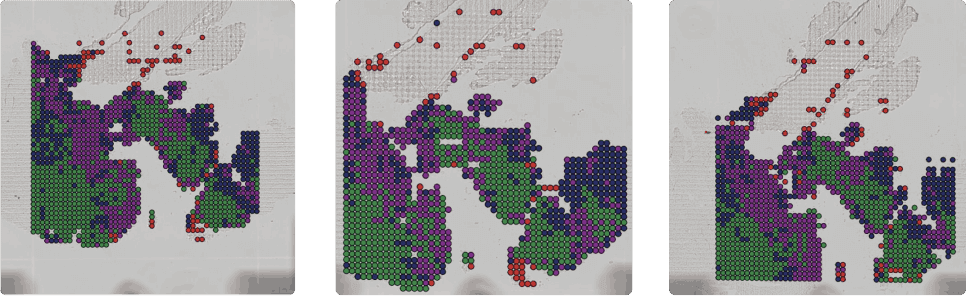
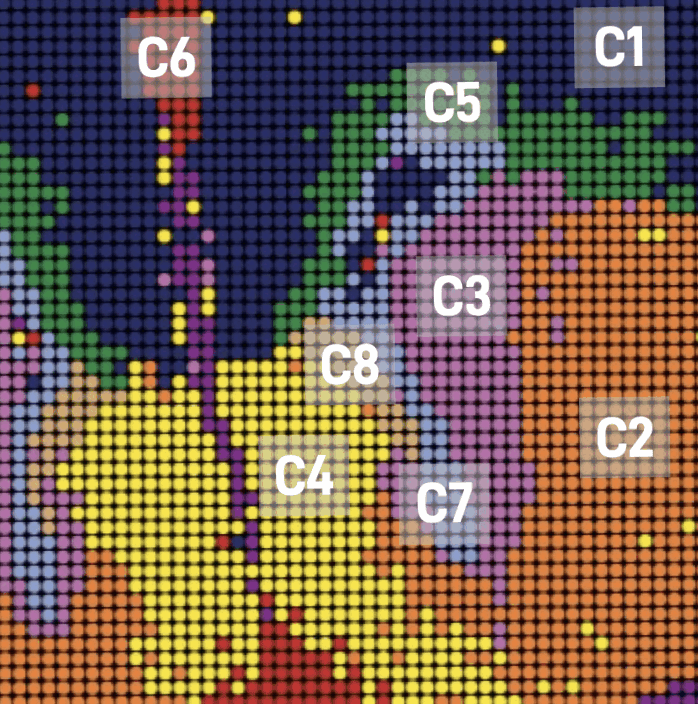
Figure 2: A tixel map of cells leading tumor infiltration (red) and cells in tumor core (green)
This information is produced in beautiful maps revealing cell identity, state, and fate in relation to epigenetic underpinnings in development and disease. With this newfound insight, you wonder why every biotech company in the world is not using this assay? This brings us to the data.
A Data Deluge
A single pair of ATAC seq files - the raw reads from a sequencing machine to produce the maps above - contains 3 billion base pairs of information. These are 30 GB in size each; imagine hundreds of files generated in a given week.
This is one of the biggest problems we have encountered. Our datasets are very large and very comprehensive, which is necessary to find unique biological insight. It can be really difficult for any users to explore the data easily.
The pipeline was previously only available through the academic lab and was hard to adopt outside of that space. You have to have all the packages set up. You need data capacity and computational infrastructure to store 100s of TBs and run 1000s of CPUs.
It’s truly difficult for anyone without expertise in bioinformatics to even look at this data... We were able to build a great pipeline internally, but in trying to get the pipeline into other people's hands, everything fell apart. Transferring the analysis pipelines and training our customers is a huge challenge to the accessibility of the product.”
The Search for Solutions
We have an awesome bioinformatics team, so first, we looked for additional bioinformaticians to help... But the size of the data required a cloud solution. So we started looking for cloud solutions to host everything, make it easy for anyone to run these tools...
Our core criteria were speed, ease of use, and ease of integration.
We talked to many different groups, many different platforms. The thing that stuck out with Latch was how quickly we were able to integrate our workflows into the platform.”
Transitioning to Latch
Despite existing infrastructure, the team decided to move their raw data, bioinformatics workflows, and downstream analysis tools to Latch. The aim was to make it easy for biologists to access, process, and visualize results.
We had already invested in other solutions and already had infrastructure set up. We had to ensure it was worth the cost of switching.

The moment that became clear - the ‘aha’ moment for us - was when all the biologists used it. It was so easy for them to adopt. They liked it, and since our customers are also biologists, we were confident that this new workflow was going to solve our problems with data transfer and training.
We talked to many different groups, many different platforms. The thing that stuck out with Latch was how quickly we were able to integrate our workflows into the platform.”
Outcomes
When we started, the analysis took 2 months. We are now nearing >2 weeks. An important part of the solution was Latch.
Today, our biologists can do very complicated analysis. This is the direction we need for customers to be able to use our platform. This is such a big accomplishment. It is a problem with spatial biology in general - these datasets are so complicated to work with. For us, making the data accessible is a huge advantage.
The flow is simple — a biologist puts in data, press run, and that’s it. She does not need to understand the script. She does not need to install packages. It’s really quite convenient. Before, this took three people.
Now, the bioinformaticians focus on the functions, the biologists focus on using it.”
Looking Forward
We are very excited about the platform, and we’re excited by how this will change the space. This enables researchers to understand their biology without the limitations of infrastructure needed to explore it.
The support from Latch has been way above expectations. It’s been great.”
The team at AtlasXomics continues to push the edge of bioinformatics to simplify access, analysis, and ease of use for their customers. To learn more about their Spatial Biology offering, go to atlasxomics.com.