Immunology
Immunoreceptor Discovery & Screening
Immunomodulation is an increasingly important strategy for combating cancer, infections, and inflammatory diseases. Fine understanding of immune cell phenotypes and T-/B-cell specificities directly translates to the creation of innovative therapeutics, such as mRNA vaccines and engineered T cells. These processes rely on TCR/BCR complex discovery and characterization. Previously a laborious multi-year endeavor, the process of finding these receptors can now be condensed into a matter of weeks using RNAseq.
Let’s explore a common use case enabled by single-cell genomics: paired ɑ/β TCR sequencing.

Derive paired ɑ/β TCR nucleotide sequences from sc-RNAseq assays
Single-cell genomics has allowed for the preservation of paired ɑ/β TCR transcripts, which would otherwise be difficult to obtain from bulk samples. Organizing TCR sequences by themselves, or with associated gene expression files, can be complex.

A bioinformatician can import all this data easily in Latch Data. And then specific TCR + GEX sequences can be organized intuitively for biologist counterparts using Latch Registry.

After all of the data has been imported, a biologist can easily find TCR + GEX sequences of interest based on functional annotations and clonotype IDs.
Align V(D)J fragments to germline and annotate clonotypes
Immunoreceptor genes are some of the most difficult to sequence on account of the biological complexity of V(D)J recombination. To begin, full-length transcripts must be obtained at the RNA level, then fragmented into V(D)J regions for alignmentment with the germline contig.
To run the analysis, a biologist can drag and drop V(D)J FastQ files, then launch cellranger vdj to process sequence fragments from 10X and generate clonotype annotations — without any code.
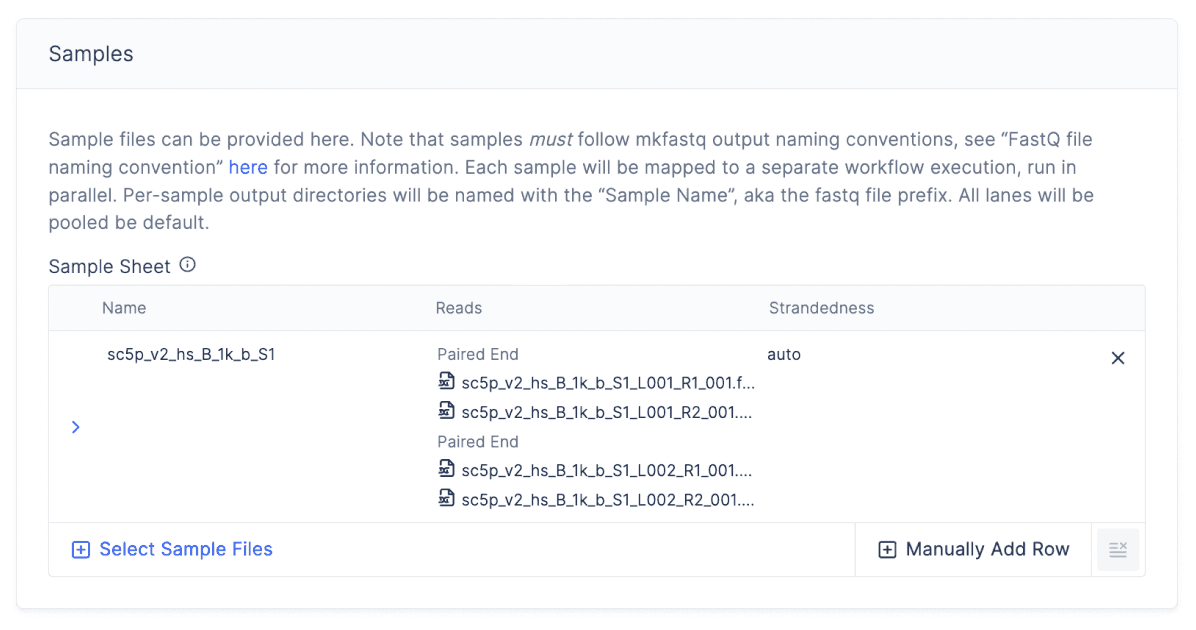
Check out the immunogenomics already available on Latch:
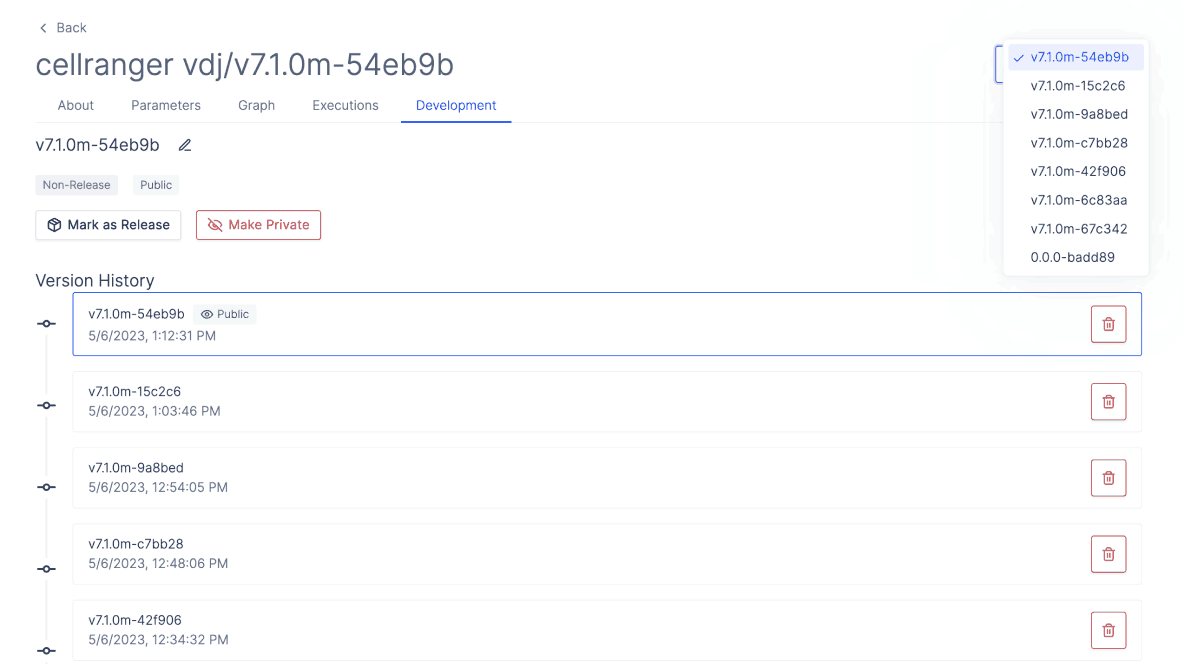
Your teams bioinformatician can also leverage the Latch SDK to upload your own custom V(D)J alignment workflow from the command line. Any workflow uploaded through the SDK allows biologists to run the workflow in a no-code GUI. It also seamlessly integrate new program updates, letting Latch auto-version your previous work.
Explore TCR clonality and generate visualizations
The immune repertoire contains incredible diversity of clonotype sequences. Observing TCR/BCR clonality over time can generate important insights into disease states and mechanisms of therapeutic response.
A biologist can explore TCR/BCR clonal dynamics using a public Pod Templates like Immunarch. View clone size and expansion kinetics over time. Extract TCR sequences of interest.
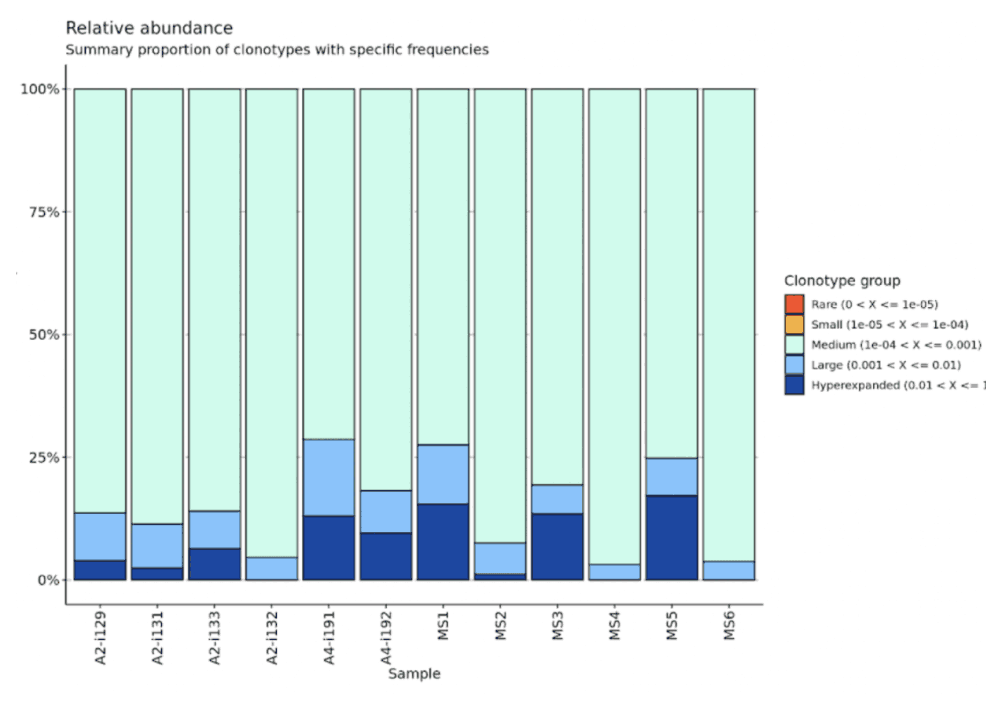
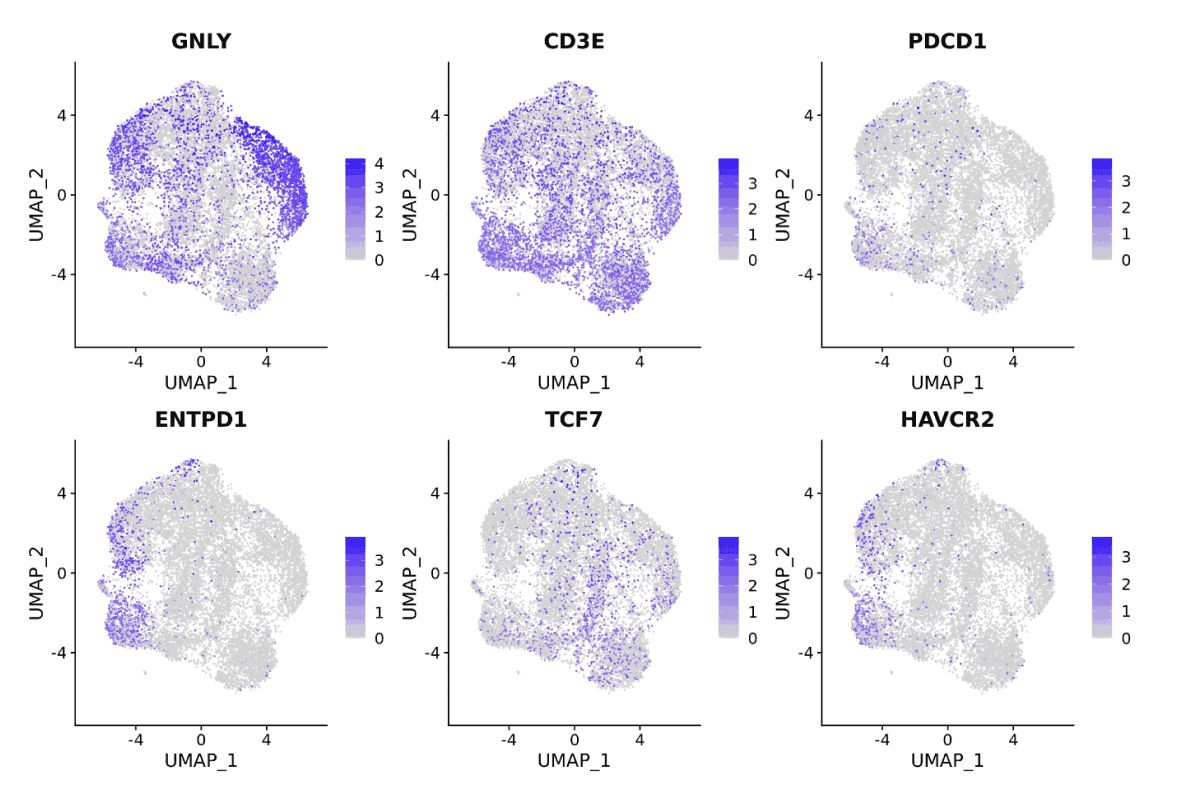
A bioinformatician can generate custom R and Python notebooks for scientist colleagues to run independently. Deliver Jupyter Notebook and R Markdown reports to collaborators using Pod Templates .
Previous
← Metagenomics